Snapshot of Peptidomics of the Red Tide Forming Species Noctiluca scintillans
- 1Guangdong Provincial Key Laboratory of Healthy and Safe Aquaculture, College of Life Science, South China Normal University, Guangzhou, China
- 2Division of Life Science, The Hong Kong University of Science and Technology, Kowloon, Hong Kong
- 3Department of Ocean Science, The Hong Kong University of Science and Technology, Kowloon, Hong Kong
- 4Key Laboratory of Marine Ecology & Environmental Sciences, Institute of Oceanology Chinese Academy of Sciences, Qingdao, China
- 5College of Marine Life Sciences, Ocean University of China, Qingdao, China
- 6Institute of Evolution and Marine Biodiversity, Ocean University of China, Qingdao, China
- 7Hong Kong Branch of Southern Marine Science & Engineering Guangdong Laboratory, The Hong Kong University of Science and Technology, Kowloon, Hong Kong
The objective of this study was to investigate the peptidome of a red tide forming species Noctiluca scintillans based on mass-spectrometry (MS) analysis. N. scintillans cells were collected at the initiation and peak phases of its bloom. Peptides were extracted by ultrafiltration with cutoff of 10 and 3 kDa in acidic condition. Optimal mass spectra were generated after purification with C18 column. MS analysis and peptide identification was undertaken by Linear Trap Quadropole (LTQ) Velos MS/MS instrument. Using this method, 239 and 332 peptide sequences from the samples collected at the initiation and peak phases of N. scintillans bloom were identified, respectively. These sequences were classified and they were affiliated to 68 and 93 groups of precursor proteins, respectively, at the initiation and peak phases, with a compositional dissimilarity of 49.5%. Meanwhile, majority of the peptides were conjectured to come from contents digested by N. scintillans. Thirty-three groups of precursor proteins were detected at both phases. These peptides involved in various intracellular physiological activities, most of them responsible for functions of digestion activities such as oxidoreductase activity, catalytic activity, hydrolase activity and cofactor/ion binding. The compositions of precursor proteins, molecular weights, functions and cell compartments of the obtained peptides were quite different between two bloom stages. This is the first attempt to study peptidomics of N. scintillans, which provide important information on the functional studies of peptidomics in N. scintillans, especially food digestion and dietary protein utilization, as well as their relationship with bloom development.
Introduction
Noctiluca scintillans is a large heterotrophic dinoflagellate (∼200–2,000 μm in diameter) that causes red tides in coastal regions worldwide (Elbrächter and Qi, 1998; Harrison et al., 2011). There are two kinds of N. scintillans, red and green (Elbrächter and Qi, 1998; Harrison et al., 2011). The N. scintillans studied herein is the red variety, without chloroplast. When no food items in cell, its cytoplasm is colorless.
The red N. scintillans is purely phagotrophic and highly omnivorous. It can feed on phytoplankton, protozoan and even the egg, larvae and adults of metazoan zooplankton (Elbrächter and Qi, 1998; Umani et al., 2004; Harrison et al., 2011; Zhang et al., 2017a). Nevertheless, phytoplankton are considered the main food items for N. scintillans in the field (Kiørboe and Titelman, 1998; Harrison et al., 2011; Zhang et al., 2017a). Previous study found that most food items in N. scintillans can be digested in about 4.5 ± 2.5 h in the field (Sang, 1991). Laboratory studies further showed that the digestion time largely depended on to the type of food N. scintillans ingest (Nikishina et al., 2011; Zhang et al., 2016, 2017c). The digestion time of Thalassiosira weissflogii was much faster than other algal food items and copepod eggs or fecal pellets (Nikishina et al., 2011; Zhang et al., 2016, 2017c). When fed on diatoms, about 90% N. scintillans cells were able to void their food vacuoles within 4 h, and 100% in 7 h (Zhang et al., 2016). Moreover, N. scintillans can survive without food for a period exceeding 3 weeks (Kiørboe and Titelman, 1998).
Peptidomics is a branch of proteomics that has been developed over the last 20 years (Zheng et al., 2006; Farrokhi et al., 2008), with the main purpose of studying transient and minute amounts of peptides and the degradome of proteases (Zheng et al., 2006; Farrokhi et al., 2008). The peptidome refers to all of the low-molecular-weight (LMW) proteins that have molecular weights less than 10 kDa (Hu et al., 2009). These peptides can be divided into two classes: (1) bioactive peptides that are produced by selective action of peptidases on larger precursor proteins, and (2) degraded peptides that result from the activity of proteolytic enzymes during protein turnover or food protein hydrolysis (Schrader and Schulz-Knappe, 2001; Adkins et al., 2002; Zheng et al., 2006; Farrokhi et al., 2008; Kadam et al., 2015). Endogenous peptides often have very specific functions as mediators and indicators of biological processes such as biologically active peptides of hormones, growth factors and cytokines, with a high impact on health and disease (Zheng et al., 2006; Farrokhi et al., 2008; Hu et al., 2009). While bioactive peptides that derived from food protein could be nutritionally beneficial as a source of essential amino acid, and exhibit various potent biological activities, e.g., antihypertensive, antioxidant, immunomodulatory and antimicrobial activities (Udenigwe and Aluko, 2012). Unfortunately, the progress in characterization of peptidomes in marine zooplankton is slow because many of these small peptides are difficult to identify and quantify due to their low abundance in cell (Geho et al., 2006; Zheng et al., 2006). Few researches studied the roles that peptidomes play in their physiological functions and intracellular biochemical processes until today (Christie, 2014a,b).
Noctiluca scintillans contains large amount of ammonia and phosphate in cell, which might be associated with the function of food digestion and buoyance (Elbrächter and Qi, 1998). It has been shown that its physiological status and the amount of intracellular ammonia and phosphate largely depend on the nutritional quality of food supply (Zhang et al., 2015, 2017b). Phytoplankton usually experience dynamic fluctuations in nutrients supply during N. scintillans bloom (Montani et al., 1998; Ara et al., 2013; Zhang et al., 2017a). However, there is no molecular level study on the relationships between the food digestion and physiological status of N. scintillans and its implication in the bloom development.
In the present study, we developed a peptidomic strategy for studying peptidome in N. scintillans at the initiation and peak phases of its bloom. The LMW peptides in N. scintillans were enriched by using filter units with a cutoff of 3 and 10 kDa, respectively. Enriched peptides were then analyzed using reverse phase high-performance liquid chromatography (RP-HPLC) and electrospray ionization tandem mass spectrometry (ESI-MS/MS) followed by strong cation exchange (SCX) chromatography without enzymatic digestion. This method allowed peptides in N. scintillans to be extracted and dissociated efficiently. A preliminary peptidomics database, with 35 positively identified precursor proteins associated with different functions of N. scintillans, was established for future physiological studies. Our study would provide foundations for future functional studies of peptidomics in microzooplankton like N. scintillans, sheds light on understanding of food digestion and dietary protein utilization in microzooplankton, as well as their relationship with bloom progression.
Materials and Methods
Sampling Collection
Samples for studying N. scintillans peptidome were collected during a field survey conducted on February 1, 2013 and February 6, 2013 in Port Shelter, a semi-enclosed bay located in the eastern part of Hong Kong (Zhang et al., 2017a). Sampling was conducted at ST 7 (22° 20.453′ N, 114° 17.703′ E), a sampling station with bottom depth of ∼17 m located in Port Shelter (Zhang et al., 2017a).
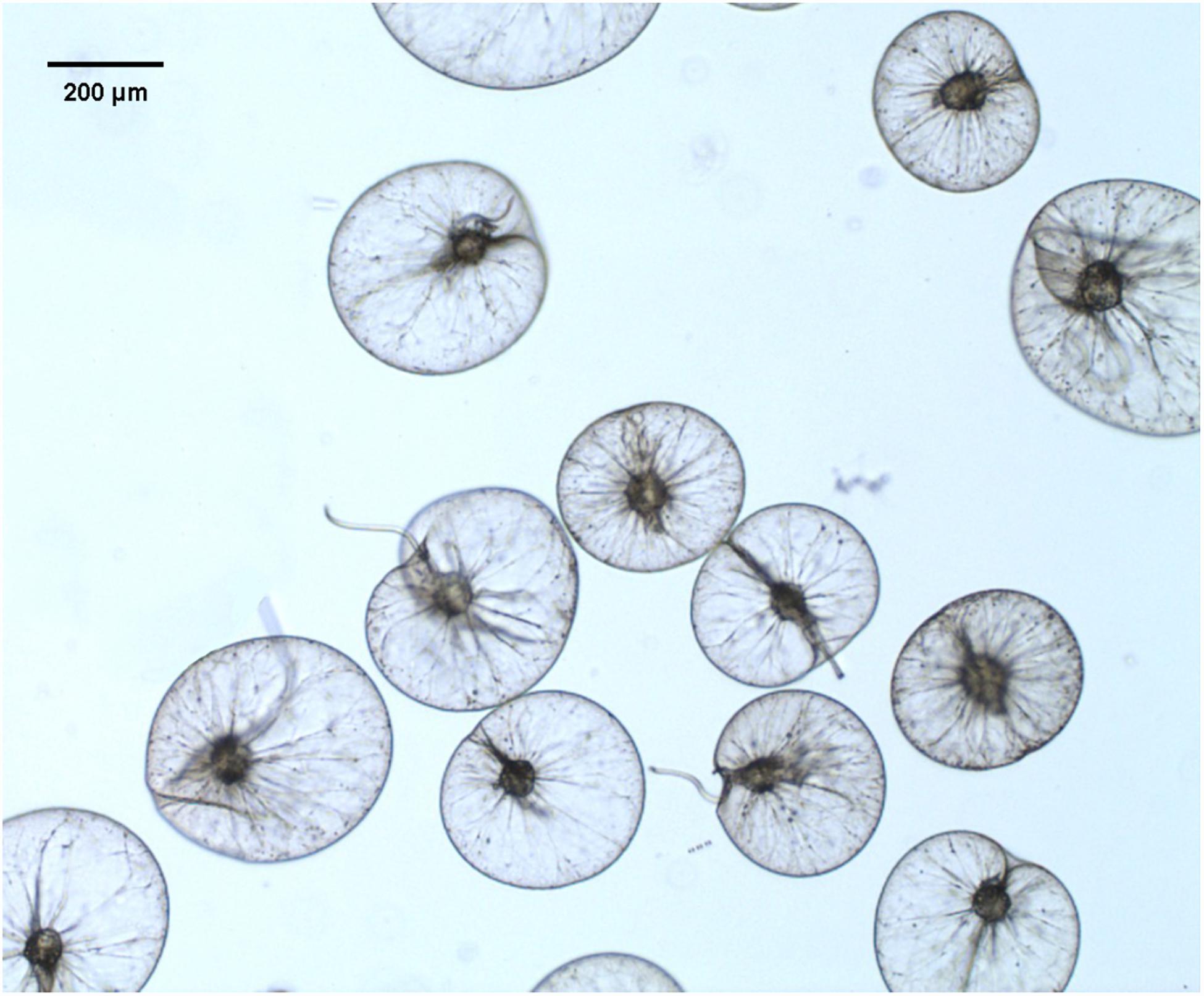
When N. scintillans bloom initiated on February 01 with concentration of 127 cells L–1 and peaked on February 06, 1976 with concentration of cells L–1, N. scintillans cells were harvested from surface seawater by using a plankton net (0.50 m diameter, 167 μm mesh size) (Zhang et al., 2017a). N. scintillans were isolated from the net-tow samples, then gently washed and resuspended with 0.2 μm-filtered autoclaved seawater. Cells were kept in the room temperature for 4 h to void food vacuoles (Sang, 1991; Zhang et al., 2016). For further purification, cells on the top surface were checked under a dissecting microscope to verify voidance of food vacuoles, and those with obvious food vacuoles were picked out. Figure 1 showed N. scintillans cells without food under the bright field. Cells were then transferred to a new beaker with autoclaved seawater and concentrated on 20-μm PC membrane. The excess seawater was absorbed by paper towels placed underneath the filter (Liu and Hastings, 2007). The cells were then transferred to 1.5-mL Eppendorf tubes and stored at −80°C until use.
Biological (size-fractionated Chl a and microplankton abundance), physical (temperature, salinity, and pH), and chemical (nutrients concentration) factors were collected and determined as described in Zhang et al. (2017a).
Peptide Extraction
Peptides were extracted from ∼6 × 105 N. scintillans cells with 1 mL of 0.25% acetic acid and methanol–water–acetic acid (90:9:1, v/v/v) solution (Baggerman et al., 2002; Husson et al., 2006; Boonen et al., 2007; Farrokhi et al., 2008). The mixtures were then shaken with vortex for 30s twice, and pipetted up and down to disrupt cells thoroughly. The extract was centrifuged at 20, 000g for 30 min, and the supernatant was concentrated using Amicon Ultra Centrifugal Filter (Millipore, United States) with a molecular weight cutoff of 10 and 3 k Da, respectively. Then the medium containing small peptides (<3 K and 3—10 K) was desalted with Sep-Pak C18 columns (Waters, Ireland) according to the manufacture’s protocol, and evaporated using speed vacuum concentrator. The peptides herein included peptides from N. scintillans itself, as well as peptides from the prey that were absorbed and utilized by N. scintillans.
SCX Fractionation of Peptide Mixture
Peptides were reconstituted and fractionated by strong cation exchange (SCX) chromatography using a protocol modified from Zhang et al. (2010). In brief, the dried peptide samples were reconstituted with 150 uL buffer A [10 mM KH2PO4, pH 3.0, acetonitrile/H2O 25/75 (v/v)] and loaded into a PolySULFOETHYL ATM column (200 mm length, 4.6 mm id, 200-Å pore size, 5 mm particle size) (PolyLC, United States) on an ÄKTA avant system (GE Healthcare, United States). The peptides were fractionated using a gradient of 100% buffer A for 5 min, 5–30% buffer B (10 mM KH2PO4, pH 3.0, 500 mM KCl, acetonitrile/H2O 25/75 (v/v)) for 40 min, 30–100% buffer B for 5 min and finally 100% buffer B for 5 min, at a constant flow rate of 1 mL min–1 for a total of 60 min. Eluted fractions were collected at 1 min interval. After combining the fractions with low peak intensity at 214 nm wavelength, a total of 5 fractions were obtained and dried using a speed vacuum. Each fraction was then reconstituted in 0.1% trifluoroacetic acid and desalted using a Sep-Pak C18 cartridge (Waters, United States). The desalted samples were dried and stored at –20°C until mass spectrometric analysis using liquid chromatography with tandem mass spectrometry (LC-MS/MS).
LC-MS/MS Analyses
Liquid chromatography with tandem mass spectrometry analyses were performed with a LTQ Velos ion trap mass spectrometer (Thermo Fisher Scientific, United States) equipped with a nano-electrospray ionization source to generate raw mass spectra data. The desalted peptide samples were dissolved in solvent A (0.1% formic acid in H2O). The nanoflow system was run at a flow rate of 500 nL min–1 with a gradient from 2 to 60% solvent B (acetonitrile with 0.1% formic acid) for 90 min and then in 80% solvent B for 2 min. The mass spectrometer was operated to automatically switch between MS and MS/MS acquisitions in a data-dependent mode. Full-scan spectra were acquired in the 400–2000 m/z range with ten most intense ions sequentially isolated for MS/MS in the linear ion trap using collision induced dissociation with dynamic exclusion.
Peptide Identification
Peak picking and deconvolution of MS/MS spectra were performed using Mascot Distiller (version 2.1.1.0) with the default parameters. Peak lists were searched against Dinophyta protein database using Mascot (version 2.2) with no enzymolysis, and following parameter settings: Methionine oxidation, C-terminal amidation, and N-terminal acetylation were set as variable modifications; peptide tolerance was set to 2 ppm, and MS/MS tolerance was 25 millimass units; the Mascot default setting of 5% was used as significance threshold. The number of incorrect target match was estimated through peptide spectrum match obtained from the decoy database, which directly enable the estimation of the well-established false discovery rate (Benjamini and Hochberg, 1995; Brosch et al., 2009). Peptides were considered identified when a score of the identity threshold corresponding to an expectation value was below 0.05.
Bioinformatic and Statistical Analyses
The obtained peptide sequences were aligned against the NCBI GenBank database by using BLAST1 to identify sequences with the highest similarity. The GI numbers of identified proteins were then imported into the searching web: http://pir.georgetown.edu/pirwww/search/batch.shtml, a protein information resource (PIR), and multiple entries were retrieved from the iProClass database. The peptides identified with the same GI numbers (i.e., same proteins) or as the subsets of same type of proteins were treated as the same precursor protein groups. For example, “heat shock protein 70” and “heat shock protein 90” were classified as “heat shock protein” (Supplementary Table 2). The Gene Ontology (GO) molecular functional and cellular component categories were then analyzed according to results of the retrieval in the iProClass database (Toulza et al., 2010; Wang et al., 2012). Compositional dissimilarities (Bray-Curtis) of the peptide molecular weight, precursors, molecular function and cellular component categories between the two samples were calculated using “vegan” in R (Oksanen et al., 2007).
Results
Environmental Factors
As shown in Table 1, N. scintillans bloom started from February 01 with concentration of 127 cells L–1, and peaked on February 06 with concentration of 1976 cells L–1. In situ environmental conditions were similar on February 01 and 06. Diatoms were abundant on February 01 (3595 cells mL–1), with Asterionella glacialis, Skeletonema costatum, T. rotula, and Pseudo-nitzschia delicatissima the dominant species. While on February 06, dinoflagellates became dominant, but with a much lower abundance (28 cell mL–1). Ciliates and mesozooplankton were also more abundant on February 06 than February 01 (Table 1; Zhang et al., 2017a). Mesodinium rubrum and Ceratium furca were the most abundant plankton species on February 06 (Supplementary Table 1).
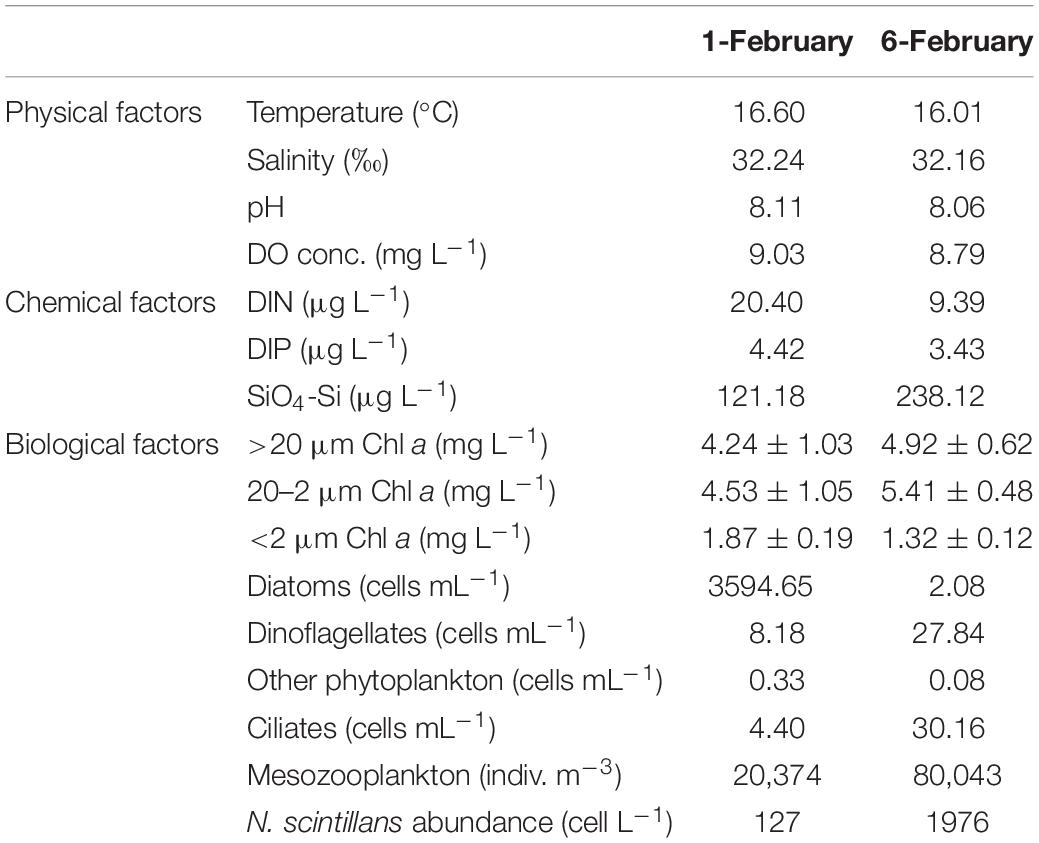
Table 1. Environmental parameters for ST7 in Port Shelter on February 01, 2013 and February 06, 2013.
Profiles of Dissociated Peptides
The samples were analyzed by LC-MS/MS after SCX fraction (Supplementary Figure 1) and reversed-phase (RP) separation. In total, 239 and 332 peptide sequences were detected at the initiation and peak phases, respectively. The majority of the peptides detected at two samples presumably arose from food digestion in N. scintillans (Figure 2). The proportions of peptides that best matched to N. scintillans were 30.13% at the initiation phase, and 8.13% at the peak phase. It was also noted that, at the initiation phase, most of the unknown peptides belonged to N. scintillans. Due to the limitation of the reference database, we only distinguished the peptides that affiliated to N. scintillans, but not explored deep into the classification of the peptides affiliated to different prey taxon in the present study.
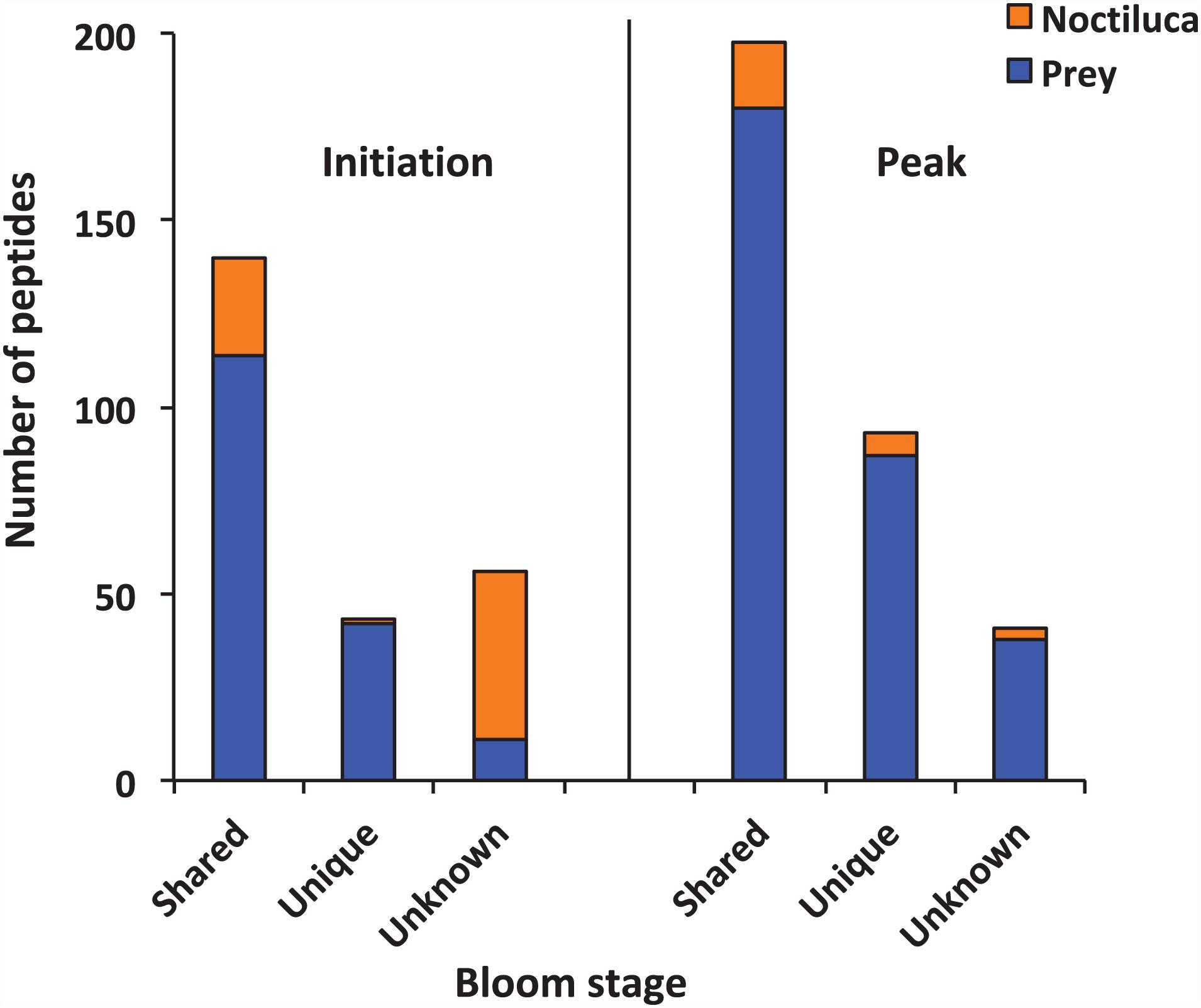
Figure 2. Numbers of peptides that were shared, unique and unknown in the samples collected at the initiation and peak phases of Noctiluca scintillans bloom.
Using BLAST, 183 sequences were classified, and they were affiliated to 68 groups of precursor proteins at the initiation phase. For the samples of the peak phase, 291 sequences were classified, and they were affiliated to 93 groups of precursor proteins (Supplementary Table 2). The compositional dissimilarity of the precursor protein groups between the two phases was 49.5% (Table 2). Thirty-three groups of precursor proteins were detected at both phases, which contributed 140 and 198 peptides at the initiation and peak phases, respectively (Figure 3A and Supplementary Table 2). The most abundant groups of precursor protein in two bloom stages were heat shock proteins (mostly heat shock protein 70 and 90), translation elongation factor and luciferase (Figure 3A and Supplementary Table 2). Meanwhile, actin and polyketide synthase at the initiation phase, and photosystem I P700 apoprotein, cytochrome c, ubiquitin conjugating enzyme, GAPDH and malate dehydrogenase at the peak phase were also abundant (>10 sequences). For those precursor proteins affiliated to N. scintillans, luciferase, heat shock protein, cysteine protease and actin were found at both phases (Figure 3B).
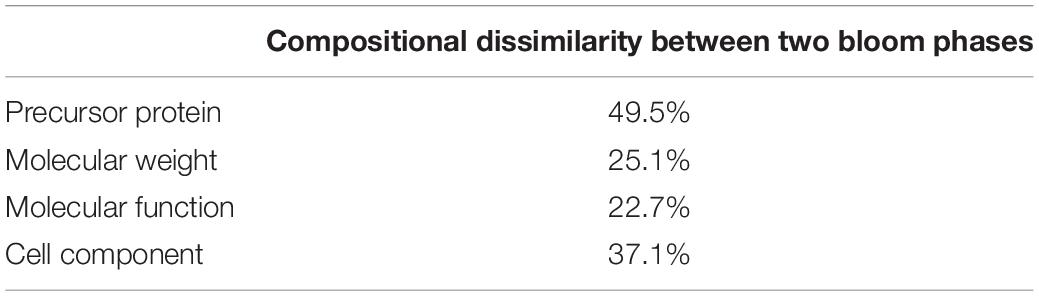
Table 2. Compositional dissimilarities of precursor protein, molecular weight of peptide, and related molecular function and cell compartment between the samples collected at the initiation and peak phases of Noctiluca scintillans bloom.
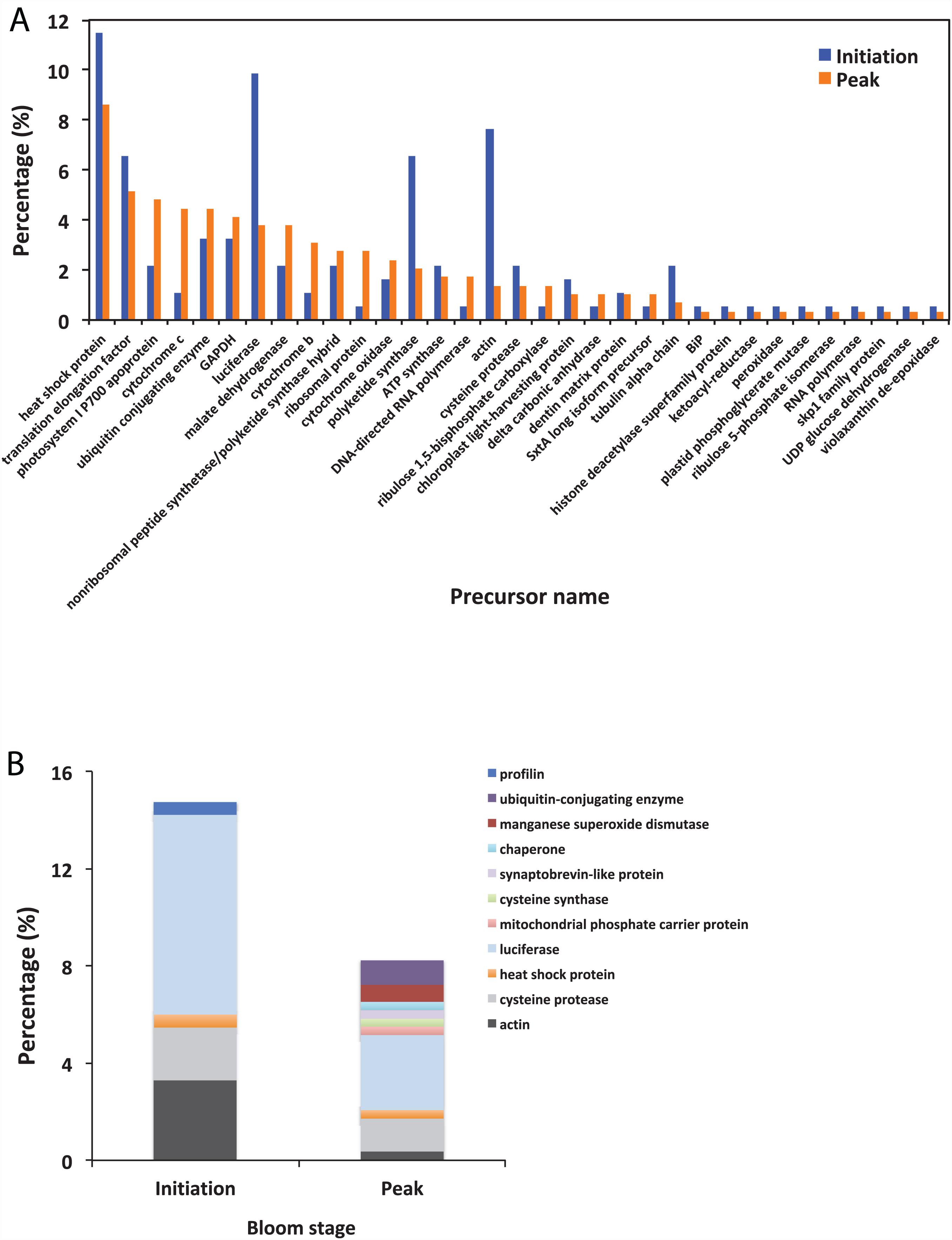
Figure 3. (A) The percentage of identified peptides sorted by shared precursor protein types in the samples collected at the initiation an peak phases of Noctiluca scintillans bloom: (B) The percentage of identified peptides affiliated to N. scintillans sorted by precursor protein groups in the samples collected at two bloom phases. Detailed information of the peptide sequences are indicated in Supplementary Table 2.
The peptides identified with a high probability assignment had molecular weight (MW) ranged 400–5,000 Da (Figure 4). Majority of them ranged from 500 to1800 Da, with 93.06% on February 01 and 94.81% on February 06. Distributions of the MW of the obtained peptides varied between initiation and peak phases (compositional dissimilarity = 25.1%; Table 2). Small peptides, whose molecular weight ranged from 500 to 1000 Da, attributed a larger proportion to the identified peptides at the peak phase (74.1%) than that at the initiation phase (49.8%). However, peptides from 1000 to 1600 Da were more abundant at the initiation phase (42.3%) than at the peak phase (20.2%). Noted that peptides between 5,000 and 10,000 Da were not significantly represented in the mass spectrometry data (data not shown). This was because that the average size of peptides of N. scintillans was 8 ± 2 residues, the charge states of some non-tryptic peptides were not high enough to be observed in the linear ion trap range of 400–2,000 m/z.
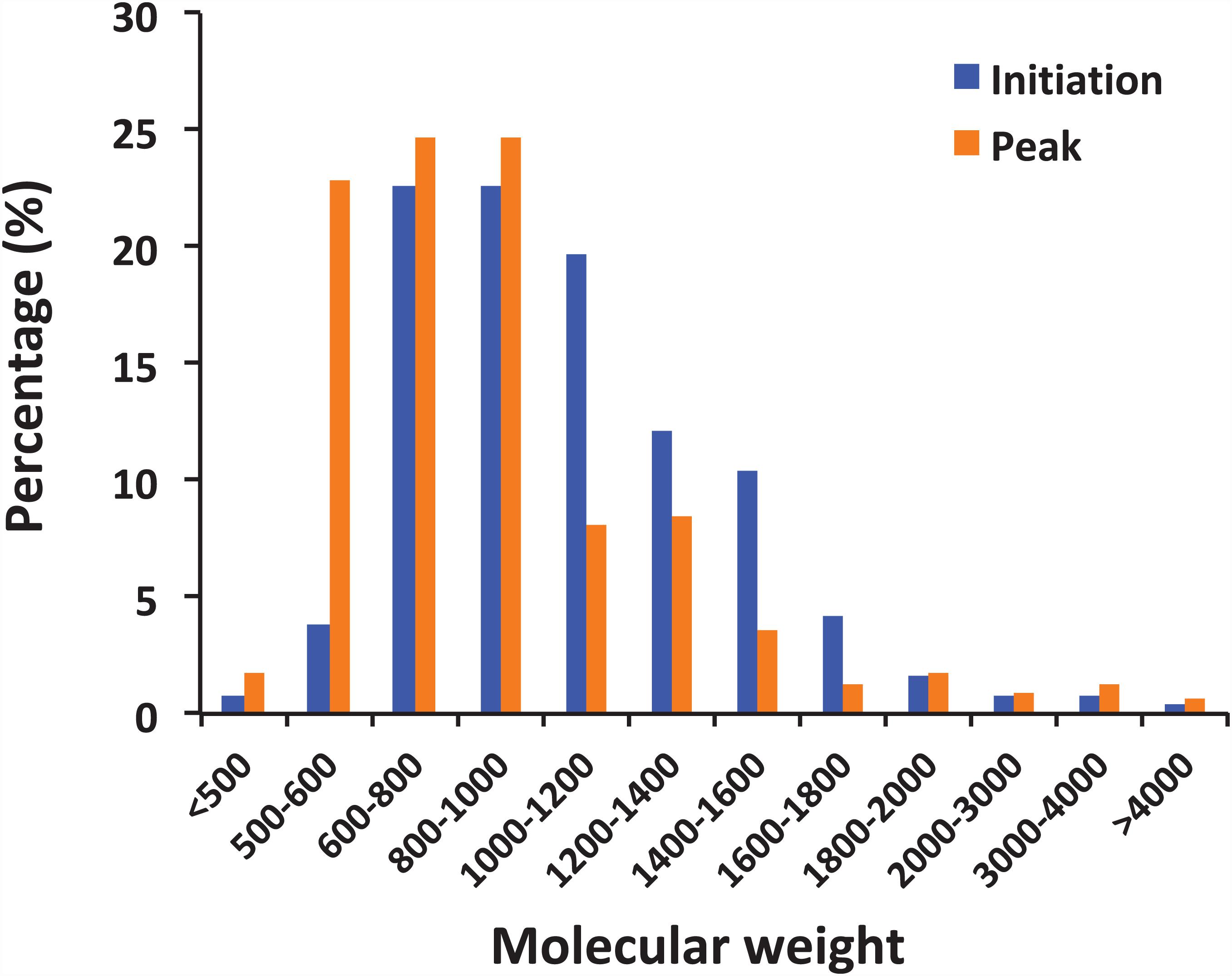
Figure 4. Distribution of peptides by molecular weight. The peptides observed in the samples from the initiation and peak phases of Noctiluca scintillans bloom with at least 2 ppm mass accuracy.
Biological Significance
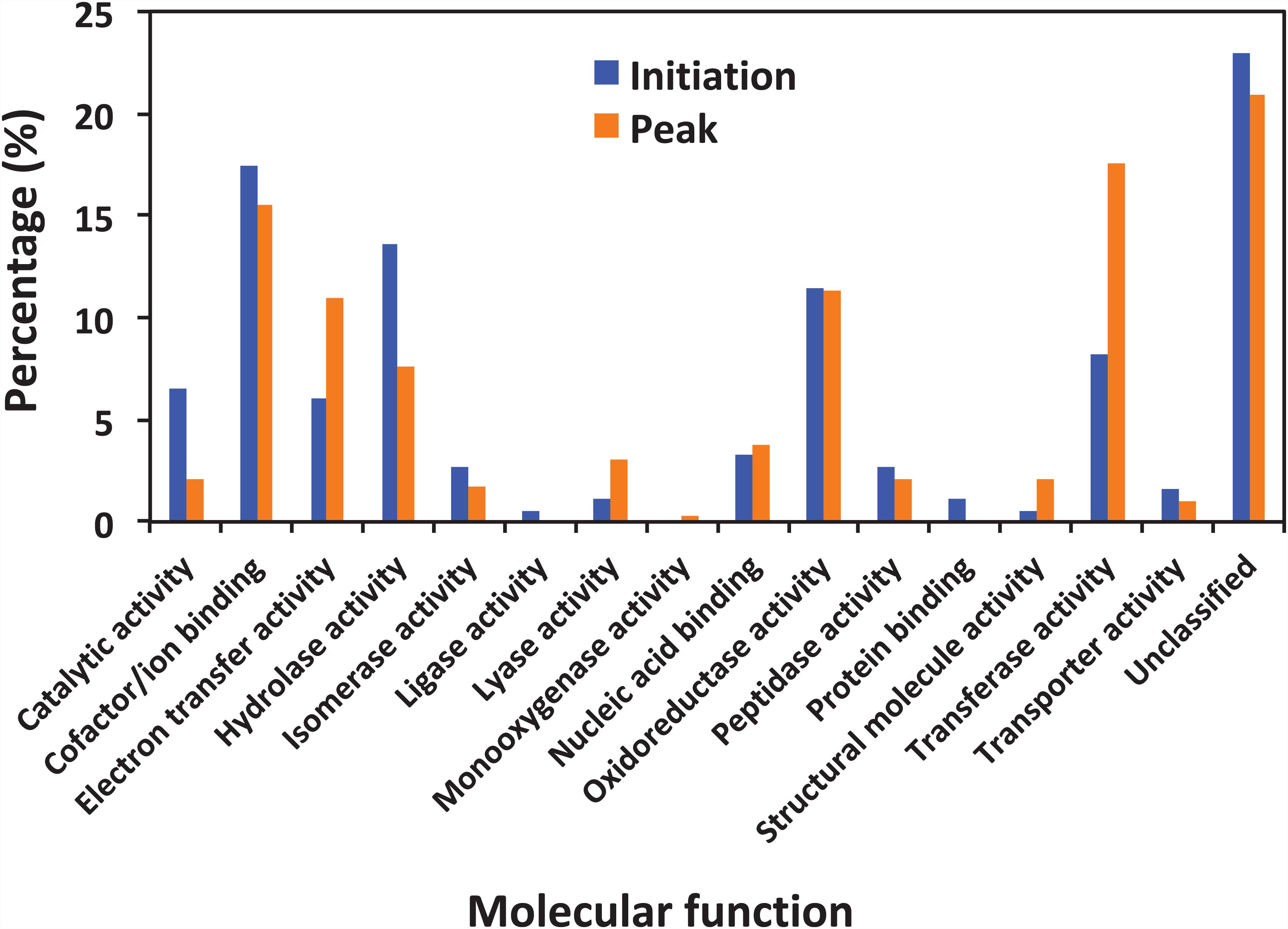
Figure 5 shows the functional classification of the precursor proteins from the samples collected at the initiation and peak phases of N. scintillans bloom based on peptide identifications. As expected, the precursor proteins were dominated by those responsible for functions of digestion activities such as oxidoreductase activity, catalytic activity, hydrolase activity and cofactor/ion binding. Others precursor groups responsible for functions like electron transfer activity, transferase activity and nucleic acid biding were also abundant (>3.0%). The compositional dissimilarity of molecular functions of the proteins of two bloom phases was 22.7% (Table 2). The cellular compartment analysis (Figure 6) showed that, chloroplast region, including plastid/thylakoid/photosystem and mitochondrion were the major compartments of the precursor proteins for both initiation and peak phases. Whereas, at the initiation phase, cytoplasm/cytoskeleton (12.0%) were a more important cell compartment than mitochondrion (4.4%).
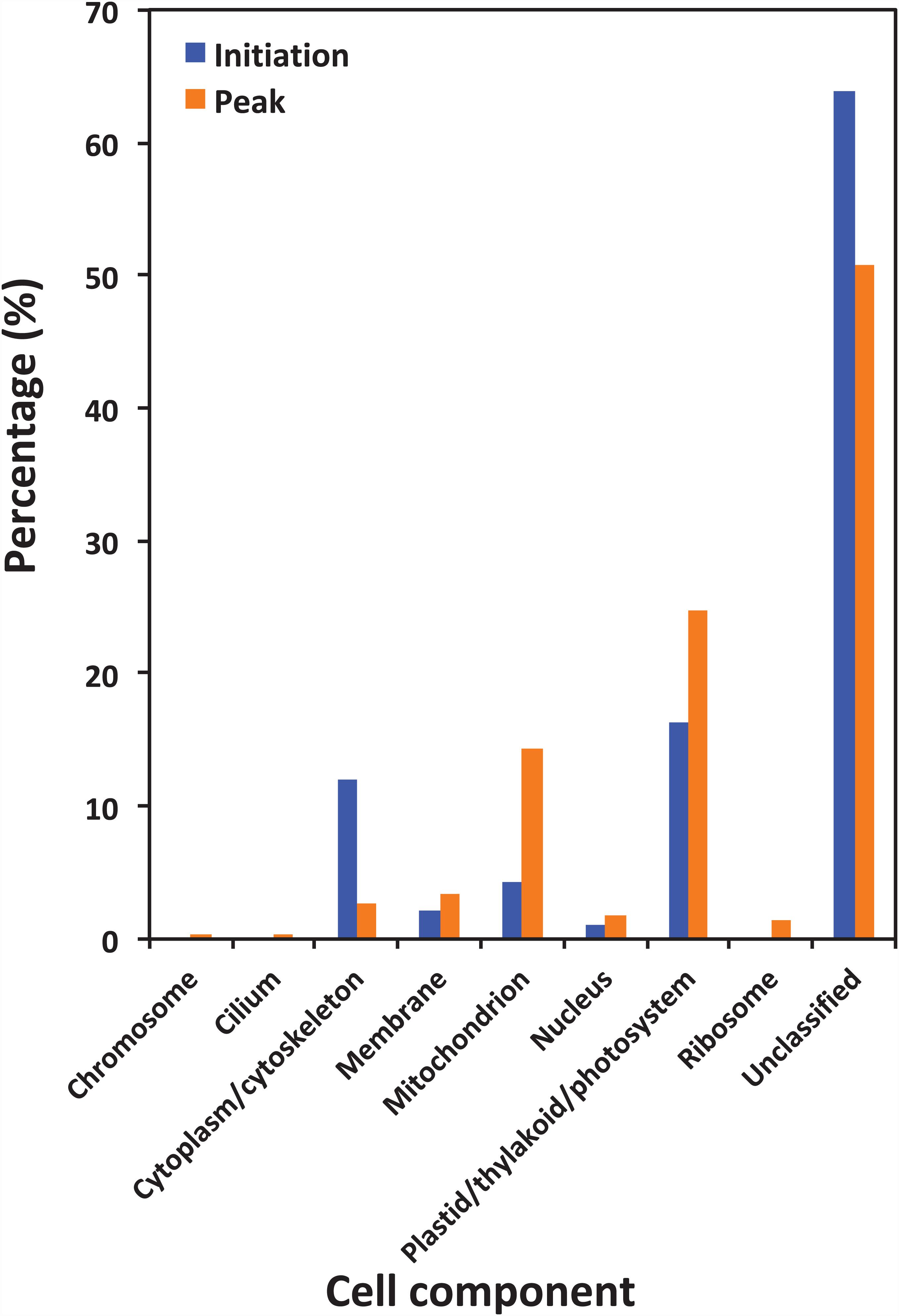
Figure 6. Gene Ontology (GO) cellular component analysis of different protein precursors between initiation and peak of Noctiluca scintillans bloom.
Discussion
A Peptidomics Approach Developed for Studying Food Digestion in N. scintillans
Peptidomics provides an effective strategy and tool for profiling and identifying physiological functions of N. scintillans, especially biochemical process of digesting and absorbing foods. Despite the potential significance of LMW peptides in biological processes (Schrader and Schulz-Knappe, 2001; Adkins et al., 2002; Zheng et al., 2006; Farrokhi et al., 2008), there was hitherto no scientific research studying peptidome in N. scintillans. In this study, we report, for the first time, the identification and characterization of the peptidomics associated with N. scintillans itself and the digested prey in it.
Identification of peptides from N. scintillans cells is difficult due to the complexity of the sample, low concentration of peptides, and difficulties in getting a protein identification from a single peptide (Zheng et al., 2006). To extract N. scintillans endogenous and food peptides, we mildly modified previous published procedures by using centrifugal ultrafiltration and by treatment without trypsin digestion (Zheng et al., 2006). Using a combination of SCX and LC-MS/MS, more than 200 unique peptides were identified, even though there are not much gene and protein information for N. scintillans and other dinoflagellate species in the public database.
Contemporary peptidomics requires prompt and confident identification of proteins of interest (Wang et al., 2011). One powerful tool for the identification of proteins from organisms with unsequenced genomes is the sequence similarity search (Shevchenko et al., 2001; Frank et al., 2005; Wang et al., 2011). In previous study, partial sequence tags or complete peptide sequences deduced directly from MS/MS spectra with no recourse to database resources were searched against a database within an error-tolerant range (Standing, 2003). Furthermore, unique peptide profiles could be used as reference maps for different parameters of specific prey quality (Onder et al., 2013). In present study, we mapped and identified the peptide fraction present in N. scintillans at two bloom stages. All things were the same, differences in the food sources were expected to lead to differences in the observed peptide profile of a given sample.
With the fast progress in sequencing technology and the increase of the available gene information for marine plankton in public database, we believed that more peptides related to microzooplankton biological processes, e.g., food digestion and dietary protein utilization will be revealed by using the approach developed in this study. Distinct peptidomic profiles detected at different bloom phases, i.e., peptide size (more 500-1,000 Da peptides at the initiation phase), proportion matching to different precursor proteins (compositional dissimilarity = 49.5%), related cell compartment (compositional dissimilarity = 37.1%), validates that this method is suitable for comparative study of the physiological functions and/or status of N. scintillans, or even other protistal microzooplankton, during different stages of bloom.
Peptidomics of N. scintillans
It is believed that many of the peptides are the fragments of larger proteins that have been at least partially degraded by various enzymes (Boire et al., 2005). For the present study, the dissociate peptides were toward self-recycling (e.g., luciferase) and food-degradation-related LMW proteins (e.g., photosystem I P700 apoprotein) that can be absorbed and utilized by N. scintillans. Therefore, the peptidomics of N. scintillans reflected a broad and balanced protein origin, including different cell compartments and exogenous soluble proteins.
Our results showed that the samples contains multiple proteases (Figure 3), many of which are vital for digestion of the prey proteins (Escalera et al., 2007; Liu and Hastings, 2007), and playing important roles in signaling and carbohydrate metabolic process as well (Figure 5). A group of precursor proteins are found associated with the function of protein degradation. For example, cysteine protease are known for their role in proteolysis of active enzymes to inactive forms, degradation of protein (Agrawal et al., 2001), and also function as peptidase. Ubiquitin-conjugating enzyme performs the second step in the ubiquitination reaction that targets a protein failed in repairing via the proteasome and lysosome (Scaglione et al., 2013). Ubiquitin and catalytic subunits of the proteasome are also expressed as precursors that are proteolytically processed to yield catalytically LMW products in eukaryotes (Varshavsky, 2012). Another example is the S-phase kinase-associated protein 1 (Skp1) family protein, a small protein and a core component of the SCF-type E3 ubiquitin ligases that mediate protein degradation by the 26S proteosome (Kong et al., 2007). Skp1 plays crucial roles in cell-cycle progression, transcriptional regulation, signal transduction, as well as many other cellular processes in eukaryotes (Hellmann and Estelle, 2002; Kong et al., 2007). Protein precursors involving in hydrolase activity include translation elongation factor-like protein and ATP synthase that catalyzes the hydrolysis of a chemical bond. Among protein lyases, although less efficient, the cyclophilin-like protein identified in this study may also have peptidyl-prolyl cis/trans-isomerase activity (Spik et al., 1991), like cyclophilin which catalyzes the isomerization of peptide bonds from trans form to cis form at proline residues and facilitates protein folding (Stamnes et al., 1992; Galat, 1993). However, proteolytic cleavage of a protein can change its solubility, charge and surface structure, especially for the hydrophobic and lipophilic membrane proteins.
In the present study, abundant precursor proteins arising from plastid/thylakoid/photosystem and mitochondrion were also identified (Figure 6). Since the N. scintillans cells tested in the present study were visibly clear of prey, this result on the one hand may reflect the rate of digestion of various prey proteins, for example, proteins in chloroplast region were slower to be digested than soluble proteins from the cytoplasm. On the other hand, the great portion of degraded peptides from plastid, mitochondrion and membrane associated proteins might indicate the preference of N. scintillans to phosphorus-rich cellular component, as those compartments are phosphorus-rich due to their phospholipid and nucleic acid chemical composition. Our previous study has shown that the growth of N. scintillans was poor when feed on phosphor-deficient algal food (Zhang et al., 2015). In addition, actively feeding N. scintillans tend to retain P and preferentially release N, highlighting its susceptible to P-limitation (Zhang et al., 2015). High P demand of N. scintillans may be due to its high DNA content like many other dinoflagellates (Rizzo, 2003; Spector, 1984). These physiological features of N. scintillans are consistent with the finding on the dominance of phosphorus-rich cellular component in the present study. It should be noted that, due to the limitation of database, the functions of ∼20% precursor proteins cannot be classified. Among them, 35.7% and 23.0% precursor proteins, respectively, at the initiation and peak phases were affiliated to N. scintillans.
It is noted that the distribution pattern of peptide size in this study is similar to the research on archaeal peptides (Kisselev et al., 1998): Peptide abundance decreases with increasing length, and less than 10% peptides exceeded 20 residues. The compositional dissimilarity of the distribution of peptide molecular weights between two bloom phases were only 25.1%. However, it is worth mentioning that small peptides, whose molecular weight ranged from 500 to 1000 Da, were more abundant at the peak phase than initiation phase (74.1% vs 49.8%; Figure 4). There are many factors that could affect the activity of food protein hydrolysates, such as specificity of proteases used for hydrolysis, degree of hydrolysis, and the structural properties of the resulting peptides including hydrophobicity, molecular size, and amino acid composition (Pihlanto, 2006; Udenigwe and Aluko, 2012). During the initiation phase, N. scintillans was observed along with a multi-species (Asterionella glacialis, Skeletonema costatum, Thalassiosira rotula, and Pseudo-nitzschia delicatissima) diatom bloom at our sampling site (Supplementary Table 1). Diatoms are believed to be the preferred food of N. scintillans (Zhang et al., 2015, 2016). At the peak phase of N. scintillans bloom, however, those diatom species were almost disappeared (Supplementary Table 1), and instead, dinoflagellates and ciliates were abundant. Based on our observation, N. scintillans did not digest food completely when food supply was sufficient, which may explain why this study detected more high molecular weight peptides at the initiation phase than at the peak phase. For animal in general, small peptides, mostly di- and tripeptides, from the small intestine are believed to be absorbed rapidly (Webb, 1990). For N. scintillans, as the bloom proceeds, the peptides become smaller in food vacuole, which leads us to speculate that food digestion of N. scintillans were more complete at the peak phase than at the initiation phase. In addition, shorter peptides would be absorbed more easily compared to those with longer residues, which supports the rapid reproduction N. scintillans and forms red tide.
Taken collectively, our study not only open a new path for future biochemical and physiological investigations of peptidoms in microzooplankton like N. scintillans, e.g., the roles played by food digestion and dietary protein utilization during different bloom stages, but also lay the groundwork for the potential development of peptide-based monitoring strategies for N. scintillans and other harmful algal blooms.
Conclusion
In this study, we made a first attempt to investigate the peptidome of N. scintillans by using modified methods for peptides extraction and identification. Approximate 200 peptides were identified, even when there are not much gene and protein information for N. scintillans and other dinoflagellate species in the public database. Distinct peptidomic profiles of N. scintillans from different bloom phases were detected, suggesting that our method is suitable for extraction, identification and comparison of peptides in N. scintillans or other marine heterotrophic planktons during different bloom stages. The functional annotation of precursor proteins and the size distribution of peptides from two bloom phases indicated that the community structure and nutrient conditions of prey might have an important effect on N. scintillans metabolism and the development of bloom. This is the first step in our endeavor to explore the relationship among N. scintillans protein/peptide size, food protein resources and reproduction. To discover the physiological and biochemical significance of peptides in N. scintillans, transcriptomic or genomic studies are need in future to provide a specific database in protein and peptide identification. With the fast progress in sequencing technology and the increase of the available gene information for marine dinoflagellates in public database, we believe that the approach presented in this study will be more useful in research on microzooplankton physiology and bloom development mechanisms.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
CL, SZ, and HL designed the study. SZ conducted the survey and collected N. scintillans. CL developed all routines for peptides extraction and identification. CL, MS, and WG enriched N. scintillans cells and conducted peptides extraction and peptides identification. SC assisted with bioinformatics analysis. CG and WG assisted with the field sampling. SZ and CL drafted the manuscript. HL and SC participated in data interpretation and modified the manuscript. All authors agreed to the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (41906117), Guangdong MEPP Fund [NO. GDOE (2019) A23]. This study was also supported by research grants from the Hong Kong Branch of Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (SMSEGL20SC01), and by the Research Grants Council of Hong Kong (T21/602/16, N_HKUST609/15, and 16128416).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to W. K. Lau for analyzing the nutrients.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2020.569807/full#supplementary-material
Supplementary Figure 1 | SCX fraction profiles of Noctiluca scintillans extracts from initiation and peak phases. (A) 3 Kda cut-off filtration. Gray curve, initiation phase; Red curve, peak phase. (B) 10 Kda cut-off filtration. Blue curve: initiation phase; Red curve: peak phase.
Supplementary Table 1 | The abundance of main plankton species in the field on February 01 and February 06, 2013.
Supplementary Table 2 | Information of the peptides, including the accession number, protein name, peptide sequence and molecular weight, molecular function and cell compartment of the peptide based on Gene Ontology, etc. The yellow filled regions were the precursor proteins found both in the initiation and peak phases. Definitions of the table headers were listed in the metadata.
Footnotes
References
Adkins, J. N., Varnum, S. M., Auberry, K. J., Moore, R. J., Angell, N. H., Smith, R. D., et al. (2002). Toward a human blood serum proteome - Analysis by multidimensional separation coupled with mass spectrometry. Mol. Cell. Proteomics 1, 947–955. doi: 10.1074/mcp.M200066-MCP200
Agrawal, M. K., Bagchi, D., and Bagchi, S. N. (2001). Acute inhibition of protease and suppression of growth in zooplankter, Moina macrocopa, by Microcystis blooms collected in Central India. Hydrobiologia 464, 37–44. doi: 10.1023/A:1013946514556
Ara, K., Nakamura, S., Takahashi, R., Shiomoto, A., and Hiromi, J. (2013). Seasonal variability of the red tide-forming heterotrophic dinoflagellate Noctiluca scintillans in the neritic area of Sagami Bay, Japan: its role in the nutrient-environment and aquatic ecosystem. Plankton Benthos Res. 8, 9–30. doi: 10.3800/pbr.8.9
Baggerman, G., Cerstiaens, A., De Loof, A., and Schoofs, L. (2002). Peptidomics of the larval Drosophila melanogaster central nervous system. J. Biol. Chem. 277, 40368–40374. doi: 10.1074/Jbc.M206257200
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate - a practical and powerful approach to multiple testing. J. R. Stat. B Methodol. 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Boire, A., Covic, L., Agarwal, A., Jacques, S., Sherifl, S., and Kuliopulos, A. (2005). PAR1 is a matrix metalloprotease-1 receptor that promotes invasion and tumorigenesis of breast cancer cells. Cell 120, 303–313. doi: 10.1016/J.Cell.2004.12.018
Boonen, K., Baggerman, G., Hertog, W. D., Husson, S. J., Overbergh, L., Mathieu, C., et al. (2007). Neuropeptides of the islets of Langerhans: a peptidomics study. Gen. Comp. Endocr. 152, 231–241. doi: 10.1016/J.Ygcen.2007.05.002
Brosch, M., Yu, L., Hubbard, T., and Choudhary, J. (2009). Accurate and sensitive peptide identification with Mascot percolator. J. Proteome Res. 8, 3176–3181. doi: 10.1021/pr800982s
Christie, A. E. (2014a). In silico characterization of the peptidome of the sea louse Caligus rogercresseyi (Crustacea, Copepoda). Gen. Comp. Endocr. 204, 248–260. doi: 10.1016/j.ygcen.2014.05.031
Christie, A. E. (2014b). Prediction of the peptidomes of Tigriopus californicus and Lepeophtheirus salmonis (Copepoda, Crustacea). Gen. Comp. Endocr. 201, 87–106. doi: 10.1016/j.ygcen.2014.02.015
Elbrächter, M., and Qi, Z. (1998). “Aspects of Noctiluca (Dinophyceae) population dynamics,” in Physiological Ecology of Harmful Algal Blooms, eds D. M. Anderson, A. D. Cembella, and G. M. Hallegraeff (Berlin: Springer), 315–335.
Escalera, L., Pazos, Y., Morono, A., and Reguera, B. (2007). Noctiluca scintillans may act as a vector of toxigenic microalgae. Harmful Algae 6, 317–320. doi: 10.1016/j.hal.2006.04.006
Farrokhi, N., Whitelegge, J. P., and Brusslan, J. A. (2008). Plant peptides and peptidomics. Plant Biotechnol. J. 6, 105–134. doi: 10.1111/j.1467-7652.2007.00315.x
Frank, A., Tanner, S., Bafna, V., and Pevzner, P. (2005). Peptide sequence tags for fast database search in mass-spectrometry. J. Proteome Res. 4, 1287–1295. doi: 10.1021/pr050011x
Galat, A. (1993). Peptidylproline cis-trans-isomerases: immunophilins. Eur. J. Biochem. 216, 689–707. doi: 10.1111/j.1432-1033.1993.tb18189.x
Geho, D. H., Liotta, L. A., Petricoin, E. F., Zhao, W. D., and Araujo, R. P. (2006). The amplified peptidome: the new treasure chest of candidate biomarkers. Curr. Opin. Chem. Biol. 10, 50–55. doi: 10.1016/j.cbpa.2006.01.008
Harrison, P., Furuya, K., Glibert, P., Xu, J., Liu, H., Yin, K., et al. (2011). Geographical distribution of red and green Noctiluca scintillans. Chin. J. Oceanol. Limnol. 29, 807–831. doi: 10.1007/s00343-011-0510-z
Hellmann, H., and Estelle, M. (2002). Plant development: regulation by protein degradation. Science 297, 793–797. doi: 10.1126/science.1072831
Hu, L., Ye, M., and Zou, H. (2009). Recent advances in mass spectrometry-based peptidome analysis. Expert Rev. Proteomic 6, 433–447. doi: 10.1586/epr.09.55
Husson, S. J., Clynen, E., Baggerman, G., Janssen, T., and Schoofs, L. (2006). Defective processing of neuropeptide precursors in Caenorhabditis elegans lacking proprotein convertase 2 (KPC-2/EGL-3): mutant analysis by mass spectrometry. J. Neurochem. 98, 1999–2012. doi: 10.1111/J.1471-4159.2006.04014.X
Kadam, S. U., Tiwari, B. K., Álvarez, C., and O’Donnell, C. P. (2015). Ultrasound applications for the extraction, identification and delivery of food proteins and bioactive peptides. Trends Food Sci. Tech. 46, 60–67. doi: 10.1016/j.tifs.2015.07.012
Kiørboe, T., and Titelman, J. (1998). Feeding, prey selection and prey encounter mechanisms in the heterotrophic dinoflagellate Noctiluca scintillans. J. Plankton Res. 20, 1615–1636. doi: 10.1093/plankt/20.8.1615
Kisselev, A. F., Akopian, T. N., and Goldberg, A. L. (1998). Range of sizes of peptide products generated during degradation of different proteins by archaeal proteasomes. J. Biol. Chem. 273, 1982–1989. doi: 10.1074/jbc.273.4.1982
Kong, H., Landherr, L. L., Frohlich, M. W., Leebens-Mack, J., Ma, H., and DePamphilis, C. W. (2007). Patterns of gene duplication in the plant SKP1 gene family in angiosperms: evidence for multiple mechanisms of rapid gene birth. Plant J. 50, 873–885. doi: 10.1111/j.1365-313X.2007.03097.x
Liu, L. Y., and Hastings, J. W. (2007). Two different domains of the luciferase gene in the heterotrophic dinoflagellate Noctiluca scintillans occur as two separate genes in photosynthetic species. Proc. Natl. Acad. Sci. U.S.A. 104, 696–701. doi: 10.1073/pnas.0607816103
Montani, S., Pithakpol, S., and Tada, K. (1998). Nutrient regeneration in coastal seas by Noctiluca scintillans, a red tide-causing dinoflagellate. J. Mar. Biotechnol. 6, 224–228.
Nikishina, A., Drits, A., Vasilyeva, Y., Timonin, A., Solovyev, K., Ratkova, T., et al. (2011). Role of the Noctiluca scintillans population in the trophic dynamics of the Black Sea plankton over the spring period. Oceanol 51, 1029–1039. doi: 10.1134/s0001437011060129
Oksanen, J., Kindt, R., Legendre, P., O’Hara, B., Stevens, M. H. H., Oksanen, M. J., et al. (2007). The vegan package. Commun. Ecolo. Package 10, 631–637.
Onder, O., Shao, W. G., Kemps, B. D., Lam, H., and Brisson, D. (2013). Identifying sources of tick blood meals using unidentified tandem mass spectral libraries. Nat. Commun. 4:1746. doi: 10.1038/Ncomms2730
Pihlanto, A. (2006). Antioxidative peptides derived from milk proteins. Int. Dairy J. 16, 1306–1314. doi: 10.1016/J.Idairyj.2006.06.005
Rizzo, P. J. (2003). Those amazing dinoflagellate chromosomes. Cell Res. 13, 215–217. doi: 10.1038/sj.cr.7290166
Sang, C. H. Q. (1991). The feeding and vegetive reproduction diurnal rhythms of Noctiluca scintillans. J. Jinan Univ. 3:022.
Scaglione, K. M., Basrur, V., Ashraf, N. S., Konen, J. R., Elenitoba-Johnson, K. S. J., Todi, S. V., et al. (2013). The ubiquitin-conjugating enzyme (E2) Ube2w ubiquitinates the N terminus of substrates. J. Biol. Chem. 288, 18784–18788. doi: 10.1074/jbc.C113.477596
Schrader, M., and Schulz-Knappe, P. (2001). Peptidomics technologies for human body fluids. Trends Biotechnol. 19, 55–60. doi: 10.1016/S0167-7799(01)01800-5
Shevchenko, A., Sunyaev, S., Loboda, A., Shevehenko, A., Bork, P., Ens, W., et al. (2001). Charting the proteomes of organisms with unsequenced genomes by MALDI-quadrupole time of flight mass spectrometry and BLAST homology searching. Anal. Chem. 73, 1917–1926. doi: 10.1021/ac0013709
Spector, D. L. (1984). Dinoflagellate Nuclei in Dinoflagellates, ed. D. L. Spector (Cambridge, MA: Academic Press), 107–147.
Spik, G., Haendler, B., Delmas, O., Mariller, C., Chamoux, M., Maes, P., et al. (1991). A novel secreted cyclophilin-like protein (Scylp). J. Biol. Chem. 266, 10735–10738.
Stamnes, M. A., Rutherford, S. L., and Zuker, C. S. (1992). Cyclophilins: a new family of proteins involved in intracellular folding. Trends Cell Biol. 2, 272–276. doi: 10.1016/0962-8924(92)90200-7
Standing, K. G. (2003). Peptide and protein de novo sequencing by mass spectrometry. Curr. Opin. Struc. Biol. 13, 595–601. doi: 10.1016/j.sbi.2003.09.005
Toulza, E., Shin, M. S., Blanc, G., Audic, S., Laabir, M., Collos, Y., et al. (2010). Gene expression in proliferating cells of the dinoflagellate Alexandrium catenella (Dinophyceae). Appl. Environ. Microb. 76, 4521–4529. doi: 10.1128/AEM.02345-09
Udenigwe, C. C., and Aluko, R. E. (2012). Food protein-derived bioactive peptides: production, processing, and potential health benefits. J. Food Sci. 77, R11–R24. doi: 10.1111/j.1750-3841.2011.02455.x
Umani, S. F., Beran, A., Parlato, S., Virgilio, D., Zollet, T., De Olazabal, A., et al. (2004). Noctiluca scintillans MACARTNEY in the Northern Adriatic Sea: long-term dynamics, relationships with temperature and eutrophication, and role in the food web. J. Plankton Res. 26, 545–561. doi: 10.1093/plankt/fbh045
Varshavsky, A. (2012). The ubiquitin system, an immense realm. Annu. Rev. Biochem. 81, 167–176. doi: 10.1146/annurev-biochem-051910-094049
Wang, D. Z., Li, C., Xie, Z. X., Dong, H. P., and Hong, H. S. (2011). Homology-driven proteomics of dinoflagellates with unsequenced genomes using MALDI-TOF/TOF and automated de novo sequencing. Evid-Based Compl. Alt. 2011, 1–16. doi: 10.1155/2011/471020
Wang, D. Z., Li, C., Zhang, Y., Wang, Y. Y., He, Z. P., Lin, L., et al. (2012). Quantitative proteomic analysis of differentially expressed proteins in the toxicity-lost mutant of Alexandrium catenella (Dinophyceae) in the exponential phase. J. Proteomics 75, 5564–5577. doi: 10.1016/j.jprot.2012.08.001
Webb, K. E. Jr. (1990). Intestinal absorption of protein hydrolysis products: a review. J. Anim. Sci. 68, 3011–3022. doi: 10.2527/1990.6893011x
Zhang, H. M., Zhao, C. Q., Li, X., Zhu, Y., Gan, C. S., Wang, Y., et al. (2010). Study of monocyte membrane proteome perturbation during lipopolysaccharide-induced tolerance using iTRAQ-based quantitative proteomic approach. Proteomics 10, 2780–2789. doi: 10.1002/pmic.201000066
Zhang, S., Harrison, P. J., Song, S., Chen, M., Kung, H. S., Lau, W. K., et al. (2017a). Population dynamics of Noctiluca scintillans during a bloom in a semi-enclosed bay in Hong Kong. Mar. Pollut. Bull. 121, 238–248. doi: 10.1016/j.marpolbul.2017.06.025
Zhang, S., Liu, H., Glibert, P. M., Guo, C., and Ke, Y. (2017b). Effects of prey of different nutrient quality on elemental nutrient budgets in Noctiluca scintillans. Sci. Rep. 7:7622. doi: 10.1038/s41598-017-05991-w
Zhang, S., Liu, H., Ke, Y., and Li, B. (2017c). Effect of the silica content of diatoms on protozoan grazing. Front. Mar. Sci. 4:202. doi: 10.3389/fmars.2017.00202
Zhang, S., Liu, H., Chen, B., and Wu, C.-J. (2015). Effects of diet nutritional quality on the growth and grazing of Noctiluca scintillans. Mar. Ecol. Prog. Ser. 527, 73–85. doi: 10.3354/meps11219
Zhang, S., Liu, H., Guo, C., and Harrison, P. J. (2016). Differential feeding and growth of Noctiluca scintillans on monospecific and mixed diets. Mar. Ecol. Prog. Ser. 549, 27–40. doi: 10.3354/meps11702
Keywords: Noctiluca scintillans, peptidomics, mass spectrometry, protein, food digestion
Citation: Zhang S, Li C, Cheung S, Sun M, Song S, Guo W, Guo C, Wu G and Liu H (2020) Snapshot of Peptidomics of the Red Tide Forming Species Noctiluca scintillans. Front. Mar. Sci. 7:569807. doi: 10.3389/fmars.2020.569807
Received: 05 June 2020; Accepted: 08 October 2020;
Published: 27 October 2020.
Edited by:
Andrew Stanley Mount, Clemson University, United StatesReviewed by:
Deana Erdner, University of Texas at Austin, United StatesChih-Ching Chung, National Taiwan Ocean University, Taiwan
Copyright © 2020 Zhang, Li, Cheung, Sun, Song, Guo, Guo, Wu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongbin Liu, liuhb@ust.hk
†These authors have contributed equally to this work
 Shuwen Zhang
Shuwen Zhang Cheng Li2†
Cheng Li2†  Cui Guo
Cui Guo Hongbin Liu
Hongbin Liu